Independent of ANY Mutation, the SARS-COVID 19 corona virus quickly proliferates as soon as the virus enters the body. In the meantime, some metabolic and chemical changes begin to occur in the body before clinical symptoms, lung symptoms and PCR test positivity have yet to occur.
In this process, the virus begins to be excreted in the urine. As the burden of the virus increases in the body, both the virus and certain substances in the urine due to metabolic changes begin to be excreted intensively.
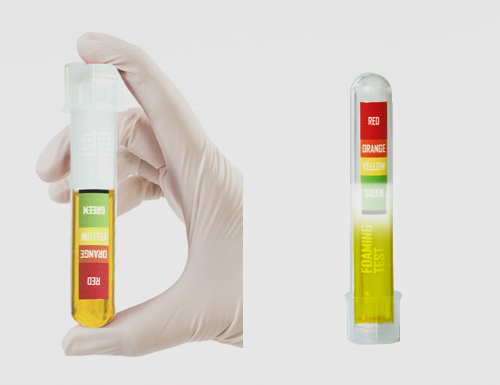
The test is conducted in a laboratory using a specially prepared reactant (CEC ) that reacts with the substances found on the surface of the virus and with the substances excreted in the urine and the sample inside the test tube foams the urine.
The rate of foaming varies in direct proportion to the virus load in the body. For people who have any viral load, less foam or more foam will be produced in the test tube, providing a VISUAL volume level indicator.
As a result of laboratory studies conducted on more than 500 patients and more than 1000 healthy individuals diagnosed with COVID-19, the amount of foam was converted into a color scale on the test tube. Validation studies have shown up to 98% correct results (Positive or Negative COVID-19 viral load).
GREEN ZONE
This indicates no viral load in the urine. Favorable clinical picture.
The green zone of the foaming test result of the patients who were ALREADY diagnosed with Covid-19 positive and HAVE ALREADY started to be treated, is interpreted as showing that the treatment process was successful and the virus load in the patient body has disappeared.
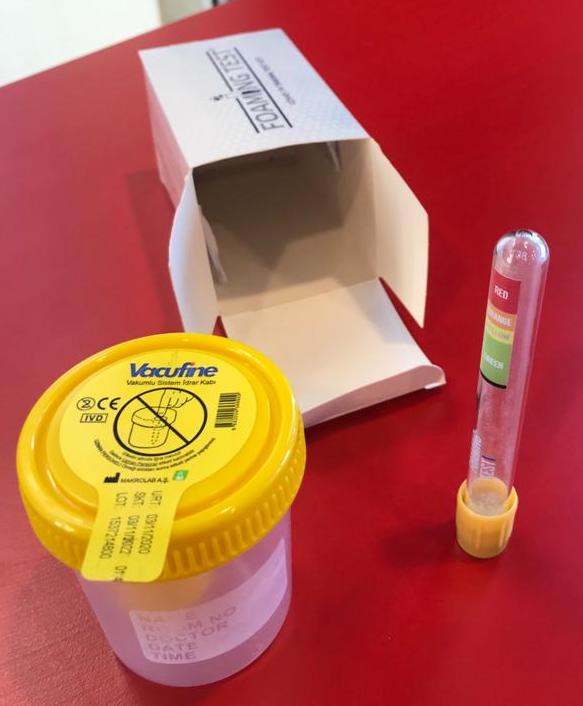
2. Fill in the test tube with Urine Sample until the black line (the Vacutainer will fill up by itself to the correct level if it was Vacuum prepared).
Anyone, anywhere can do all stages of the test under hygienic conditions. Depending on your location and Test Result, conserve or dispose of the Samples in a Safe Manner. ALWAYS Follow local Health Department regulations. NOTE: The 98% Probability Diagnostics is the result of Validation testing, available to Customers and Research Organisations. Here is the FULL PROCEDURE:

About our PCR testing kits: Our ORSAIR PhoenixDX 2019-NCOV is a real-time RT-PCR-based detection system for the 2019 Wuhan coronavirus (2019-nCoV). 2019-nCoV is considered a novel human coronavirus that is genetically distinct from the common human coronaviruses (229E, NL63, OC43, HKU1), which cause seasonal acute respiratory illness. It is also genetically distinct from the two newer human coronaviruses, MERS-CoV and SARS-CoV.
Our Test Kit (50 tests) ORSAIR PhoenixDX 2019-NCOV detects the presence of 3 different and highly specific gene sequences of 2019-nCoV: E gene, N gene and RdRP gene. All 3 assays must be tested positive to confirm the sample as 2019-nCoV-positive.
Additionally, a non-infectious positive control and a negative human extraction control are included. The positive control is used to confirm functionality of the assays and overall PCR performance, the negative human extraction control is included to evaluate the quality of the RNA isolation independently from the 2019-nCoV assays.The first step in the detection of 2019-nCoV is the conversion of viral RNA into cDNA. Afterwards, the target sequences unique for 2019-nCoV are specifically amplified with amplification monitored in real time through the use of fluorescently labelled probes: upon incorporation into the newly amplified DNA strands, the fluorophore (FAMTM) is released and an increase in fluorescence signal can be observed.
Due to the intrinsic mutation rate of coronaviruses, it is possible that mutations in the target sequence occur and accumulate over time. This can lead to false-negative results with a PCR- based detection approach. ORSAIR PhoenixDX 2019-NCOV addresses this issue by using 3 detection assays on 3 different target sequences to minimize the chance of false-negative results caused by an altered target sequence.
If samples are tested negative in one or more assays, additional complementary testing may be required. The original target sequences for 2019-nCoV are included as a non-infectious target positive control (TPC) to check the integrity of the detection assays.
Samples tested positive should always be confirmed through complementary methods and additional analysis in an independent laboratory.
3.3) SPECIMENS - HANDLING AND STORAGE
Specimens can be stored at 4°C for up to 72 hours after collection.
If a delay in extraction is expected, store specimens at -70°C or lower.
Extracted nucleic acids should be stored at -70°C or lower.
Do not use specimens if:
they were not kept at 2-4°C (≤ 4 days) or frozen at -70°C or below.
they are insufficiently labelled or lack documentation.
they are not suitable for this purpose (see above for suitable sample material).
the specimen volume is insufficient.
3.4) SAMPLE PREPARATION / NUCLEIC ACID EXTRACTION
The performance of RT-PCR assays strongly depends on the amount and quality of sample template RNA. It is strongly recommended to qualify and validate RNA extraction procedures for recovery and purity before testing specimens.
original prada bags anguish (1987) gucci website usa chatgpt vs gpt3 carlos prada brown wig cap shop gucci prada canapa bag plus followers instagram re edition prada prada little bag edward tian chatgpt raffia prada bag prada nylon sneakers weird spiky fruit prada jeans women real captains hat prada nylon skirt 5000 instagram followers chatgpt 免费 velvet prada bag men prada necklace gucci shopping online microsoft chatgpt investment self addressed stamped postcard recent instagram followers gratis followers instagram prada blanket gucci outlet bags brown goatee prada jackets christian dior outlet online lisa golightly hazy beach 50 followers instagram bag prada jazz festival catalina island vancouver riots kissing couple prada bifold wallet tack boards chatgpt 代理 prada socks mens prada cropped jacket chatgpt api github prada dog bandana gucci outlet bags prada slip ons jack spade darrow brief adodas outlet prada messenger vintage prada dress gg clothing brand prada dallas yachting cap women gucci com prada cologne macy's in the ghetto lisa marie and elvis itunes uses for chatgpt pantone indigo blue bing chatgpt waitlist analyze followers instagram red prada sandals beard size chart jeff goldblum biz markie prada sunglasses discount vancouver riots kissing aaatravel.com prada law chatgpt slow chatgpt powerpoint reebok outlet online englewood ymca fruit with spiky outside prada luna rose dan command chatgpt gucci fashion house prada ski suit prada black tie prada ostrich bag chatgpt homework prada cleo white prada buckle belt pr17ws prada chatgpt client www mix co prada au prada tenis hombre mnii prada backpack large prada mask red fruit with spikes bondy bait online clothing outlet stores prada pr 57xs купить гуччи prada puffer boots how to make a fake mustache with makeup prada reedition 2005 prada crossbody men rambutan nutrition prada painting make wigs igfamed.com instagram followers green prada wallet tods shoes outlet electric mashman snowboard helmet puma premium outlets prada zip gucci hk
Suitable nucleic acid extraction systems successfully used in combination with ORSAIR PhoenixDX DETECTION KITS include: bioMérieux NucliSens® systems, QIAamp® Viral RNA Mini Kit, QIAamp® MinElute Virus Spin Kit or RNeasy® Mini Kit (QIAGEN), EZ1 DSP Virus Kit (QIAGEN), Roche MagNA Pure Compact RNA Isolation Kit, Roche MagNA Pure Compact Nucleic Acid Isolation Kit, and Roche MagNA Pure 96 DNA and Viral NA Small Volume Kit, and Invitrogen ChargeSwitch® Total RNA Cell Kit.
Store and keep residual specimens and extracted nucleic acids at -70°C.
Only thaw the number of specimen extracts that will be tested in a single day. Do not freeze/thaw extracts more than once before testing as each freeze/thaw cycle will decrease the RNA quality.
It may be possible to use patient samples directly, depending on the sample type. However, this may require a prior lysis step and titration of the amount on sample that can be used without inhibiting the reaction. This procedure has not been validated, use of isolated RNA is recommended.
Please email for more information and a ask QUOTE = always indicate if you are a End-User or not and WHICH MODEL of PCR are your using? Mr.Theo BASCH, theo.orsys@gmail.com